Using Biopython and matplotlib would seem like the way to go, indeed.
It really just boils down to three lines of code to get that graph:
import Bio, pandas
lengths = map(len, Bio.SeqIO.parse('/path/to/the/seqs.fasta', 'fasta'))
pandas.Series(lengths).hist(color='gray', bins=1000)
Of course you might want to make a longer script that's callable from the command line, with a couple options. You are welcome to use mine:
#!/usr/bin/env python2
"""
A custom made script to plot the distribution of lengths
in a fasta file.
Written by Lucas Sinclair.
Kopimi.
You can use this script from the shell like this:
$ ./fastq_length_hist --input seqs.fasta --out seqs.pdf
"""
###############################################################################
# Modules #
import argparse, sys, time, getpass, locale
from argparse import RawTextHelpFormatter
from Bio import SeqIO
import pandas
# Matplotlib #
import matplotlib
matplotlib.use('Agg', warn=False)
from matplotlib import pyplot
################################################################################
desc = "fasta_length_hist v1.0"
parser = argparse.ArgumentParser(description=desc, formatter_class=RawTextHelpFormatter)
# All the required arguments #
parser.add_argument("--input", help="The fasta file to process", type=str)
parser.add_argument("--out", type=str)
# All the optional arguments #
parser.add_argument("--x_log", default=True, type=bool)
parser.add_argument("--y_log", default=True, type=bool)
# Parse it #
args = parser.parse_args()
input_path = args.input
output_path = args.out
x_log = bool(args.x_log)
y_log = bool(args.y_log)
################################################################################
# Read #
lengths = map(len, SeqIO.parse(input_path, 'fasta'))
# Report #
sys.stderr.write("Read all lengths (%i sequences)\n" % len(lengths))
sys.stderr.write("Longest sequence: %i bp\n" % max(lengths))
sys.stderr.write("Shortest sequence: %i bp\n" % min(lengths))
sys.stderr.write("Making graph...\n")
# Data #
values = pandas.Series(lengths)
# Plot #
fig = pyplot.figure()
axes = values.hist(color='gray', bins=1000)
fig = pyplot.gcf()
title = 'Distribution of sequence lengths'
axes.set_title(title)
axes.set_xlabel('Number of nucleotides in sequence')
axes.set_ylabel('Number of sequences with this length')
axes.xaxis.grid(False)
# Log #
if x_log: axes.set_yscale('symlog')
if y_log: axes.set_xscale('symlog')
# Adjust #
width=18.0; height=10.0; bottom=0.1; top=0.93; left=0.07; right=0.98
fig.set_figwidth(width)
fig.set_figheight(height)
fig.subplots_adjust(hspace=0.0, bottom=bottom, top=top, left=left, right=right)
# Data and source #
fig.text(0.99, 0.98, time.asctime(), horizontalalignment='right')
fig.text(0.01, 0.98, 'user: ' + getpass.getuser(), horizontalalignment='left')
# Nice digit grouping #
sep = ('x','y')
if 'x' in sep:
locale.setlocale(locale.LC_ALL, '')
seperate = lambda x,pos: locale.format("%d", x, grouping=True)
axes.xaxis.set_major_formatter(matplotlib.ticker.FuncFormatter(seperate))
if 'y' in sep:
locale.setlocale(locale.LC_ALL, '')
seperate = lambda x,pos: locale.format("%d", x, grouping=True)
axes.yaxis.set_major_formatter(matplotlib.ticker.FuncFormatter(seperate))
# Save it #
fig.savefig(output_path, format='pdf')
EDIT - an example output:
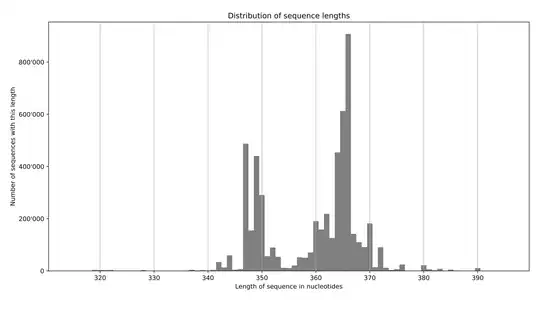
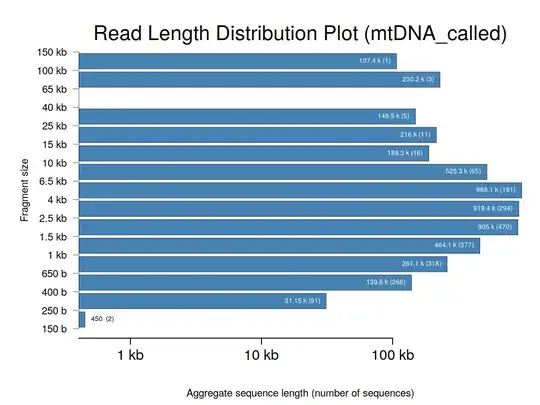


faidx --transform chromsizes | cut -f2 | Rscript -e 'data <- as.numeric (readLines ("stdin")); summary(data); hist(data)'. This requires pyfaidx:pip install pyfaidx. – Matt Shirley May 26 '17 at 13:35