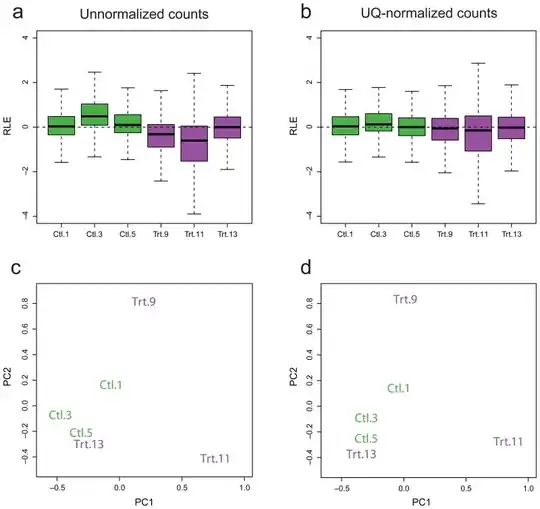
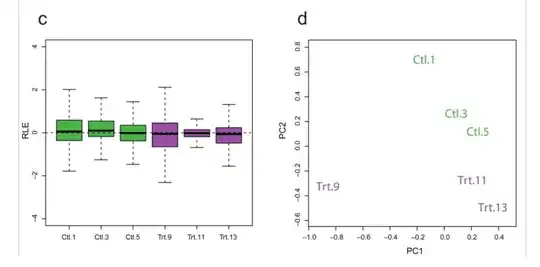
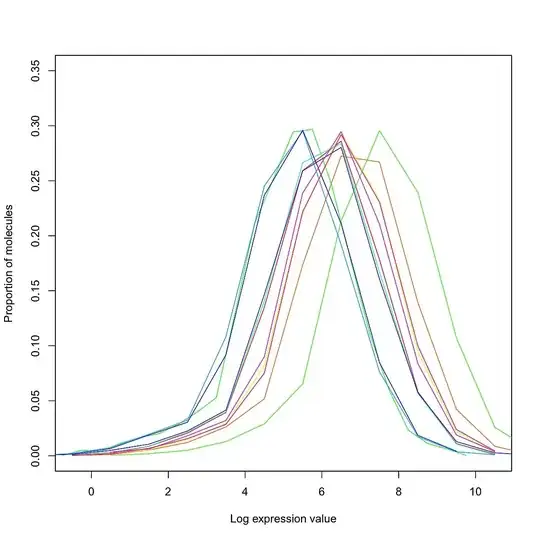
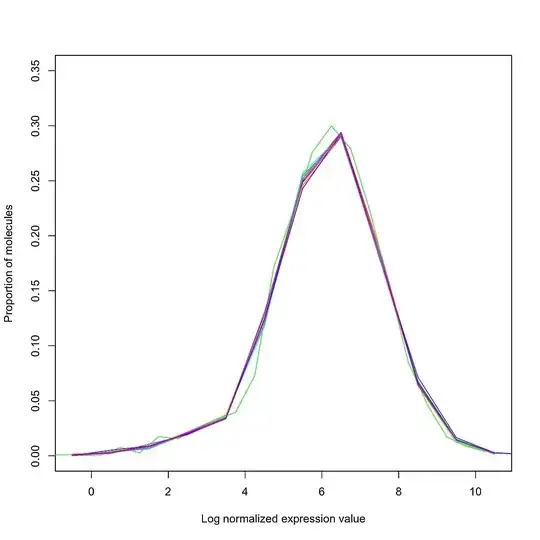
I am working with a set of (bulk) RNA-Seq data collected across multiple runs, run at different times of the year. I have normalized my data using library size / quantile / RUV normalization, and would like to check (quantitatively and/or qualitatively) whether or not normalization has succeeded in removing the batch effects.
It is important to note that by "normalization has succeeded", I simply mean that unwanted variation has been removed - further analysis is required to ensure that biological variation has not been removed. What are some plots / statistical tests / software packages which provide a first-step QC for normalization?